Generic Model of Eukaryotic Cell Cycle Control: Running Simulations
Coupled with the JigCell Comparator, the JigCell Run Manager allows a user to specify and run an ensemble of simulations.
Before using the Run Manager, the user first runs facilitator.exe (located in directory JigCell/facilitator), which allows the simulator to talk with the Run Manager, then runs xppserver.bat (in directory JigCell) which starts the simulator).
The Run Manager uses a spreadsheet interface. There are two tabs (labeled as Runs and Settings, respectively).
In each row of the Runs spreadsheet, as shown in Fig. 8, the user specifies how to simulate an experiment, giving the names for the model equations (i.e., the name of the SBML file), the parameter values (the name of individual run), the initial conditions (the name of the initial conditions for that run) and the the numerical algorithm to use (the name of the integrator). After specifying these names, right clicking on the row and selecting the Plot option will cause a graph with simulation results to appear.
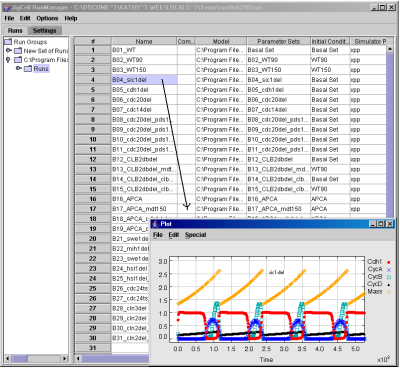
Fig. 8. The Runs spreadsheet summarizes the conditions for simulations for the wild type and mutant cells in the budding yeast.
The information for each name that appears in the columns of the Runs spreadsheet is defined in the three Settings spreadsheets.
- Parameters, Fig. 9, contains a list of parameter settings for each simulation run. The information in this spreadsheet is stored in 3 separate files: model.sbml.basp (for the basal parameter set), model.sbml.basi (for the basal initial conditions), and model.sbml.pm (a name list of simulation runs and their corresponding parameter changes).
- Initial Conditions, Fig. 10, contains a name list of initial conditions and their changes. This information is stored in the file model.sbml.ic.
- Simulation Parameters, Fig. 11, contains a list of integrators. This information, together with the path of the sbml file and the setting files (model.sbml, model.sbml.basp, model.sbml.basi, model.sbml.pm, model.sbml.ic) are stored in the run file itself, in model.sbml.run. Note that in JigCell version 6.0.0, the absolute path is used; it will be revised and changed to the relative path in a future release.
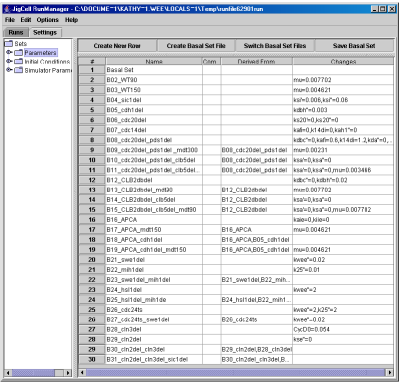
Fig. 9. The Parameters Settings spreadsheet defines the parameter values for each simulation.
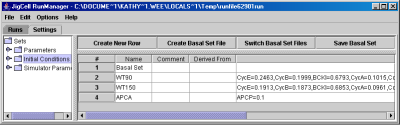
Fig. 10. The Initial Conditions Settings spreadsheet.
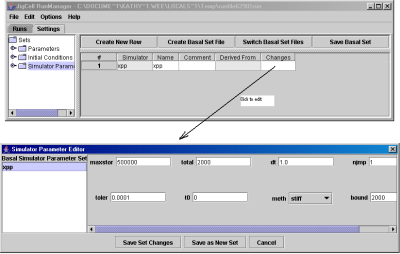
Fig. 11. The Simulation Parameters Settings spreadsheet.
To create a new run file, first the user needs to define the SBML file to be used (to create the basal parameter set and the basal initial conditions set from it), and to define simulator settings. Fig. 12 shows how to create the basal parameter set from the SBML file. Fig. 13 illustrates how to define the parameter changes for a particular simulation run.
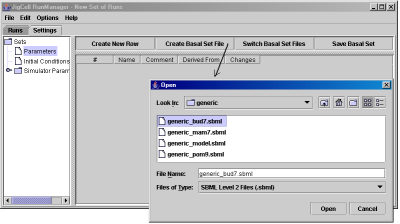
Fig. 12. Create the basal parameter set from the model SBML file.
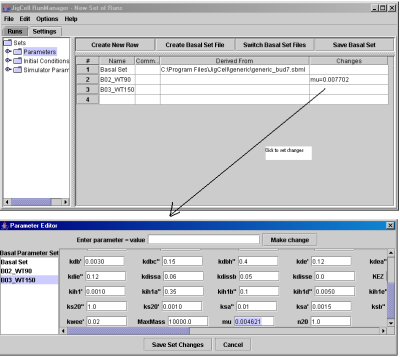
Fig. 13. Add the parameter setting for a particular simulation.
Once the Parameter Settings spreadsheet contains all the settngs (one for each simulation), and the spreadsheets for Initial Conditions Settings and Simulation Parameter Settings are completed, then the user can define individual runs in the Runs spreadsheet as shown in Fig. 8.
Click on the links to see screen shots of the Run Manager files for budding yeast, fission yeast, and mammalian cells.
By organizing information in a table, the Run Manager allows the modeler to explore parameter space easily by doing batch runs over a complicated set of simulations.